Explodable 3D Dog Skull for Veterinary Education
3D models of a Sheep and Goat Skull and Inner ear
3D models of Miocene vertebrates from Tavers
3D GM dataset of bird skeletal variation
Skeletal embryonic development in the catshark
Bony connexions of the petrosal bone of extant hippos
bony labyrinth (11) , inner ear (10) , Eocene (8) , South America (8) , Paleobiogeography (7) , skull (7) , phylogeny (6)
Lionel Hautier (23) , Maëva Judith Orliac (21) , Laurent Marivaux (16) , Rodolphe Tabuce (14) , Bastien Mennecart (13) , Pierre-Olivier Antoine (12) , Renaud Lebrun (11)

|
Virtual reconstruction of a Late Jurassic metriorhynchid skull from Switzerland and its use for scientific illustration and paleoartSophie De Sousa Oliveira, Léa Girard
Published online: 19/07/2023 |

|
M3#1037Left dentary (3 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1037 state:published |
Download 3D surface file |

|
M3#1038Right dentary (3 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1038 state:published |
Download 3D surface file |

|
M3#1039Left ramus (2 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1039 state:published |
Download 3D surface file |

|
M3#1040Right ramus (3 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1040 state:published |
Download 3D surface file |

|
M3#1041Left splenial (2 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1041 state:published |
Download 3D surface file |

|
M3#1042Right splenial (2 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1042 state:published |
Download 3D surface file |

|
M3#1043Frontal and left prefrontal Type: "3D_surfaces"doi: 10.18563/m3.sf.1043 state:published |
Download 3D surface file |

|
M3#1044Left maxilla (4 meshes) Type: "3D_surfaces"doi: 10.18563/m3.sf.1044 state:published |
Download 3D surface file |

|
M3#1045Right maxilla Type: "3D_surfaces"doi: 10.18563/m3.sf.1045 state:published |
Download 3D surface file |

|
M3#1046Left nasal Type: "3D_surfaces"doi: 10.18563/m3.sf.1046 state:published |
Download 3D surface file |

|
M3#1047Right nasal Type: "3D_surfaces"doi: 10.18563/m3.sf.1047 state:published |
Download 3D surface file |

|
M3#1048Parietal Type: "3D_surfaces"doi: 10.18563/m3.sf.1048 state:published |
Download 3D surface file |

|
M3#1049Right postorbital Type: "3D_surfaces"doi: 10.18563/m3.sf.1049 state:published |
Download 3D surface file |

|
M3#1050Right prefrontal Type: "3D_surfaces"doi: 10.18563/m3.sf.1050 state:published |
Download 3D surface file |

|
M3#1051Right premaxilla Type: "3D_surfaces"doi: 10.18563/m3.sf.1051 state:published |
Download 3D surface file |

|
M3#1052Left squamosal Type: "3D_surfaces"doi: 10.18563/m3.sf.1052 state:published |
Download 3D surface file |

|
M3#1053Right squamosal Type: "3D_surfaces"doi: 10.18563/m3.sf.1053 state:published |
Download 3D surface file |

|
M3#1054Reconstruction of the mandible Type: "3D_surfaces"doi: 10.18563/m3.sf.1054 state:published |
Download 3D surface file |

|
M3#1055Reconstruction of the cranium Type: "3D_surfaces"doi: 10.18563/m3.sf.1055 state:published |
Download 3D surface file |

This contribution contains the three-dimensional digital models of the dental fossil material of strepsirrhine primates (Azibiidae and ?Djebelemuridae) from the late early to early middle Eocene of the Gour Lazib Complex in western Algeria and of Djebel Chambi in central-western Tunisia. These fossils were described, figured and discussed in the following publication: Marivaux et al. (2025), New insights into the diversity of strepsirrhine primates from the late early – early middle Eocene of North Africa (Algeria and Tunisia). Journal of Human Evolution, 103729. https://doi.org/10.1016/j.jhevol.2025.103729
Algeripithecus minimissimus ONM-CBI-1-38 View specimen

|
M3#1715Isolated right P3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1715 state:published |
Download 3D surface file |
Algeripithecus minimissimus ONM-CBI-1-37 View specimen

|
M3#1716Isolated right P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1716 state:published |
Download 3D surface file |
Algeripithecus minimissimus ONM-CBI-1-1206 View specimen

|
M3#1717Isolated right p4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1717 state:published |
Download 3D surface file |
Algeripithecus minimissimus ONM-CBI-1-1207 View specimen

|
M3#1718Isolated right p4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1718 state:published |
Download 3D surface file |
Algeripithecus minimissimus ONM-CBI-1-1205 View specimen

|
M3#1719Fragment of right mandible bearing m1-3 (Holotype) Type: "3D_surfaces"doi: 10.18563/m3.sf.1719 state:published |
Download 3D surface file |
Algeripithecus minimissimus ONM-CBI-1-1209 View specimen

|
M3#1720Isolated left m2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1720 state:published |
Download 3D surface file |
Algeripithecus minimissimus ONM-CBI-1-1208 View specimen

|
M3#1721Isolated right m2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1721 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-294 View specimen

|
M3#1722Left DP4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1722 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-297 View specimen

|
M3#1723Isolated right P2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1723 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-298 View specimen

|
M3#1724Isolated right P3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1724 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-299 View specimen

|
M3#1725Isolated right P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1725 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-303 View specimen

|
M3#1726Isolated left P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1726 state:published |
Download 3D surface file |
Algeripithecus minutus UM-GZC-7 View specimen

|
M3#1727Isolated left M1 (lingually broken) Type: "3D_surfaces"doi: 10.18563/m3.sf.1727 state:published |
Download 3D surface file |
Algeripithecus minutus UM-GZC-1 View specimen

|
M3#1728Isolated left M2 (Holotype) Type: "3D_surfaces"doi: 10.18563/m3.sf.1728 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-319 View specimen

|
M3#1729Isolated left M3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1729 state:published |
Download 3D surface file |
Algeripithecus minutus UM-HGL50-397 View specimen

|
M3#1730Fragment of left mandible bearing p3-m3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1730 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-258 View specimen

|
M3#1731Isolated right P3 or P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1731 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-260 View specimen

|
M3#1732Isolated right M2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1732 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-261 View specimen

|
M3#1733Isolated left M3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1733 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-263 View specimen

|
M3#1734Isolated left p3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1734 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-264 View specimen

|
M3#1735Isolated right m1 (Holotype) Type: "3D_surfaces"doi: 10.18563/m3.sf.1735 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-265 View specimen

|
M3#1736Isolated right m1 (lingually broken) Type: "3D_surfaces"doi: 10.18563/m3.sf.1736 state:published |
Download 3D surface file |
Azibius magnus UM-HGL50-266 View specimen

|
M3#1738Isolated right m2 (corroded) Type: "3D_surfaces"doi: 10.18563/m3.sf.1738 state:published |
Download 3D surface file |
Azibius trerki UM-HGL50-166 View specimen

|
M3#1739Isolated right DP4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1739 state:published |
Download 3D surface file |
Azibius trerki UM-HGL50-295 View specimen

|
M3#1740Isolated left DP4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1740 state:published |
Download 3D surface file |
Azibius trerki UM-HGL51-46 View specimen

|
M3#1741Fragment of right maxillary bearing P3-4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1741 state:published |
Download 3D surface file |

|
M3#1742Fragment of right maxillary bearing M3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1742 state:published |
Download 3D surface file |
Azibius trerki UM-GZC-41 View specimen

|
M3#1743Isolated left P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1743 state:published |
Download 3D surface file |
Azibius trerki UM-HGL50-396 View specimen

|
M3#1744Boneless fragment of a left maxillary bearing M1-2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1744 state:published |
Download 3D surface file |
Azibius trerki UM-HGL50-270 View specimen

|
M3#1745Fragment (talonid) of an isolated right dp4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1745 state:published |
Download 3D surface file |
Azibius trerki UM-HGL50-248 View specimen

|
M3#1746Isolated left m1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1746 state:published |
Download 3D surface file |
Azibius trerki UM-HGL50-256 View specimen

|
M3#1753Fragment of left mandible bearing p4-m3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1753 state:published |
Download 3D surface file |
Lazibadapis anchomomyinopsis UM-HGL50-326 View specimen

|
M3#1747Isolated right M1 (buccally broken) Type: "3D_surfaces"doi: 10.18563/m3.sf.1747 state:published |
Download 3D surface file |
Lazibadapis anchomomyinopsis UM-HGL50-169 View specimen

|
M3#1748Isolated right M2 (corroded) Type: "3D_surfaces"doi: 10.18563/m3.sf.1748 state:published |
Download 3D surface file |
Lazibadapis anchomomyinopsis UM-HGL50-170 View specimen

|
M3#1749Isolated right M2 or M3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1749 state:published |
Download 3D surface file |
Lazibadapis anchomomyinopsis UM-HGL50-325 View specimen

|
M3#1750Boneless fragment of left mandible preserving m2-3 (Holotype) -> m2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1750 state:published |
Download 3D surface file |

|
M3#1751Boneless fragment of left mandible preserving m2-3 (Holotype) -> m3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1751 state:published |
Download 3D surface file |
Lazibadapis anchomomyinopsis UM-HGL50-290 View specimen

|
M3#1752Isolated left m3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1752 state:published |
Download 3D surface file |
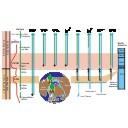
This contribution contains the 3D models described and figured in the following publication: Tabuce R., Marandat B., Adnet S., Gernelle K., Girard F., Marivaux L., Solé F., Schnyder J., Steurbaut E., Storme J.-Y., Vianey-Liaud M., Yans J. (2025). European mammal turnover driven by a global rapid warming event preceding the Paleocene-Eocene Thermal Maximum. PNAS. https://doi.org/10.1073/pnas.2505795122
Acritoparamys aff. atavus UM-ALB-41 View specimen

|
M3#17653D digital model Type: "3D_surfaces"doi: 10.18563/m3.sf.1765 state:published |
Download 3D surface file |
Acritoparamys aff. atavus UM-ALB-42 View specimen

|
M3#1766m1 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1766 state:published |
Download 3D surface file |
Acritoparamys aff. atavus UM-ALB-43 View specimen

|
M3#1767M3 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1767 state:published |
Download 3D surface file |
indet. indet. UM-ALB-7 View specimen

|
M3#1768M1or2 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1768 state:published |
Download 3D surface file |
Arcius cf. rougieri UM-ALB-3 View specimen

|
M3#1769m2 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1769 state:published |
Download 3D surface file |
Arfia sp. UM-ALB-2 View specimen

|
M3#1770M1or2 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1770 state:published |
Download 3D surface file |
Bustylus sp. UM-ALB-37 View specimen

|
M3#1771M1 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1771 state:published |
Download 3D surface file |
?Corbarimys sp. UM-ALB-44 View specimen

|
M3#1772M1or2 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1772 state:published |
Download 3D surface file |
indet. indet. UM-ALB-26 View specimen

|
M3#1773upper molar (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1773 state:published |
Download 3D surface file |
indet. indet. UM-ALB-39 View specimen

|
M3#1774m1or2 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1774 state:published |
Download 3D surface file |
Paschatherium marianae UM-ALB-4 View specimen

|
M3#1775P4 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1775 state:published |
Download 3D surface file |
Paschatherium marianae UM-ALB-5 View specimen

|
M3#1776DP4 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1776 state:published |
Download 3D surface file |
Paschatherium marianae UM-ALB-8 View specimen

|
M3#1777mandible with m2 and talonid of m1 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1777 state:published |
Download 3D surface file |
Paschatherium marianae UM-ALB-10 View specimen

|
M3#1778M3 (righ Type: "3D_surfaces"doi: 10.18563/m3.sf.1778 state:published |
Download 3D surface file |
Paschatherium marianae UM-ALB-22 View specimen

|
M3#1779m3 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1779 state:published |
Download 3D surface file |
Paschatherium marianae UM-ALB-33 View specimen

|
M3#1780M2 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1780 state:published |
Download 3D surface file |
Peratherium sp. UM-ALB-12 View specimen

|
M3#1781?m2 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1781 state:published |
Download 3D surface file |
Peratherium sp. UM-ALB-23 View specimen

|
M3#1782?M2 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1782 state:published |
Download 3D surface file |
Peratherium sp. UM-ALB-25 View specimen

|
M3#1783?M3 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1783 state:published |
Download 3D surface file |
Plagioctenodon cf. dormaalensis UM-ALB-16 View specimen

|
M3#1784M1or2 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1784 state:published |
Download 3D surface file |
Plagioctenodon cf. dormaalensis UM-ALB-18 View specimen

|
M3#1785P4 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1785 state:published |
Download 3D surface file |
gen. nov. sp. nov. UM-ALB-27 View specimen

|
M3#1786M1or2 (left) Type: "3D_surfaces"doi: 10.18563/m3.sf.1786 state:published |
Download 3D surface file |
Teilhardimys cf. reisi UM-ALB-36a View specimen

|
M3#1787M2 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1787 state:published |
Download 3D surface file |
Teilhardimys cf. reisi UM-ALB-36b View specimen

|
M3#1788M1 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1788 state:published |
Download 3D surface file |
Wyonycteris sp. UM-ALB-19 View specimen

|
M3#1789M1or2 (right) Type: "3D_surfaces"doi: 10.18563/m3.sf.1789 state:published |
Download 3D surface file |

Speothos pacivorus is an extinct South American canid (Canidae: Cerdocyonina) from the Pleistocene of Lagoa Santa Karst, Central Brazil. This taxon is one of the hypercarnivore canids that vanished from the continent at the end of Pleistocene. Although all remains of Speothos pacivorus were collected in the 19th century by the Danish naturalist Peter W. Lund, few studies have committed to an in-depth analysis of the taxon and the known specimens. Here, we analyzed all biological remains of S. pacivorus hosted in the Peter Lund/Quaternary Collection at the Natural History Museum of Denmark, Copenhagen, by listing and illustrating all its specimens known to date. We also conducted a reconstruction of the holotype, an almost complete cranium, based on a µCT scan, producing an undeformed and crack-free three-dimensional model. With this data available we aim to foster new research on this elusive species.
Speothos pacivorus NHMD:211341 View specimen

|
M3#1475Holotype of Speothos pacivorus Type: "3D_surfaces"doi: 10.18563/m3.sf.1475 state:published |
Download 3D surface file |

We provide a 3D reconstruction of the skull of Latimeria chalumnae that can be easily accessed and visualized for a better understanding of its cranial anatomy. Different skeletal elements are saved as separate PLY files that can be combined to visualize the entire skull or isolated to virtually dissect the skull. We included some guidelines for a fast and easy visualization of the 3D skull.
Latimeria chalumnae MHNG 1080.070 View specimen
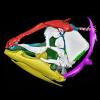
|
M3#1254the skeletal elements of the skull of Latimeria chalumnae included in 26 different PLY files Type: "3D_surfaces"doi: 10.18563/m3.sf.1254 state:published |
Download 3D surface file |
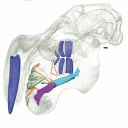
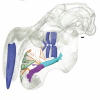
This contribution contains the 3D models described and figured in the following publication: Hautier L, Gomes Rodrigues H, Ferreira-Cardoso S, Emerling CA, Porcher M-L, Asher R, Portela Miguez R, Delsuc F. 2023. From teeth to pad: tooth loss and development of keratinous structures in sirenians. Proceedings of the Royal Society B. https://doi.org/10.1098/rspb.2023.1932
Dugong dugon 2005.51 View specimen
|
M3#1275Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1275 state:published |
Download 3D surface file |
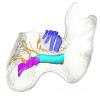
Dugong dugon 2023.66 View specimen
|
M3#1274Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1274 state:published |
Download 3D surface file |
Dugong dugon 5386 View specimen

|
M3#1276Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1276 state:published |
Download 3D surface file |
Dugong dugon 1848.8.29.7/GERM 1027g View specimen

|
M3#1277Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1277 state:published |
Download 3D surface file |
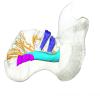
Dugong dugon 1991.413 View specimen
|
M3#1278Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1278 state:published |
Download 3D surface file |
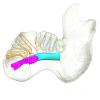
Dugong dugon 1991.427 View specimen
|
M3#1279Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1279 state:published |
Download 3D surface file |
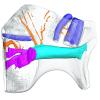
Dugong dugon 2017-3-9 View specimen
|
M3#1280Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1280 state:published |
Download 3D surface file |
Eosiren lybica 1913-22 View specimen

|
M3#1281Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1281 state:published |
Download 3D surface file |
Halitherium taulannense RGHP C001 View specimen

|
M3#1282Internal mandibular morphology. Cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1282 state:published |
Download 3D surface file |
Halitherium taulannense RGHP C009 View specimen
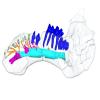
|
M3#1283Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1283 state:published |
Download 3D surface file |
Hydrodamalis gigas 1947.10.21.1 View specimen
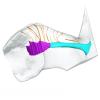
|
M3#1284Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal Type: "3D_surfaces"doi: 10.18563/m3.sf.1284 state:published |
Download 3D surface file |
Hydrodamalis gigas C1021 View specimen

|
M3#1285Anterior part of the mandible Type: "3D_surfaces"doi: 10.18563/m3.sf.1285 state:published |
Download 3D surface file |

|
M3#1286Posterior part of the mandible Type: "3D_surfaces"doi: 10.18563/m3.sf.1286 state:published |
Download 3D surface file |
Hydrodamalis gigas 2023.67 View specimen
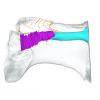
|
M3#1287Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal Type: "3D_surfaces"doi: 10.18563/m3.sf.1287 state:published |
Download 3D surface file |
Libysiren sickenbergi M.82429 View specimen

|
M3#1288Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1288 state:published |
Download 3D surface file |
Libysiren sickenbergi M.45675 View specimen

|
M3#1289Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1289 state:published |
Download 3D surface file |
Prorastomus sirenoides OR.448976 View specimen

|
M3#1290Internal morphology of the left mandible. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1290 state:published |
Download 3D surface file |

|
M3#1304Internal morphology of the right mandible. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1304 state:published |
Download 3D surface file |
Ribodon limbatus M.7073 View specimen
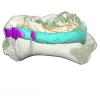
|
M3#1292Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1292 state:published |
Download 3D surface file |
Rytiodus capgrandi PAL2017-8-1 View specimen

|
M3#1293Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1293 state:published |
Download 3D surface file |
Trichechus inunguis 1868.12.19.2 View specimen
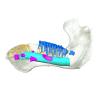
|
M3#1294Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1294 state:published |
Download 3D surface file |
Trichechus manatus 1843.3.10.12 View specimen

|
M3#1295Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1295 state:published |
Download 3D surface file |
Trichechus manatus 1864.6.5.1 View specimen
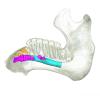
|
M3#1296Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1296 state:published |
Download 3D surface file |
Trichechus manatus 1950.1.23.1 View specimen

|
M3#1297Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1297 state:published |
Download 3D surface file |
Trichechus senegalensis 1885.6.30.2 View specimen
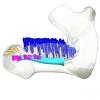
|
M3#1298Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1298 state:published |
Download 3D surface file |
Trichechus senegalensis 1894.7.25.8 View specimen

|
M3#1299Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1299 state:published |
Download 3D surface file |
Trichechus senegalensis V97 View specimen
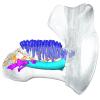
|
M3#1302Mandibular internal morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1302 state:published |
Download 3D surface file |
Trichechus sp. 65.4.28.9 View specimen

|
M3#1300Internal mandibular morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth; green = tooth alveoli. Type: "3D_surfaces"doi: 10.18563/m3.sf.1300 state:published |
Download 3D surface file |
Dugong dugon 1946.8.6.2 View specimen

|
M3#1301Mandibular internal morphology. Orange = dorsal canaliculi; purple = mental branches; cyan = mandibular canal; dark blue = teeth. Type: "3D_surfaces"doi: 10.18563/m3.sf.1301 state:published |
Download 3D surface file |

This contribution contains 3D models of left and right house mouse (Mus musculus domesticus) inner ears analyzed in Renaud et al. (2024). The studied mice belong to four groups: wild-trapped mice, wild-derived lab offspring, a typical laboratory strain (Swiss) and hybrids between wild-derived and Swiss mice. They have been analyzed to assess the impact of mobility reduction on inner ear morphology, including patterns of divergence, levels of inter-individual variance (disparity) and intra-individual variance (fluctuating asymmetry)
Mus musculus Tourch_7819 View specimen

|
M3#1366Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1366 state:published |
Download 3D surface file |
Mus musculus Tourch_7821 View specimen

|
M3#1367Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1367 state:published |
Download 3D surface file |
Mus musculus Tourch_7839 View specimen

|
M3#1368Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1368 state:published |
Download 3D surface file |
Mus musculus Tourch_7873 View specimen

|
M3#1369Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1369 state:published |
Download 3D surface file |
Mus musculus Tourch_7877 View specimen

|
M3#1370Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1370 state:published |
Download 3D surface file |
Mus musculus Tourch_7922 View specimen

|
M3#1371Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1371 state:published |
Download 3D surface file |
Mus musculus Tourch_7923 View specimen

|
M3#1372Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1372 state:published |
Download 3D surface file |
Mus musculus Tourch_7925 View specimen

|
M3#1373Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1373 state:published |
Download 3D surface file |
Mus musculus Tourch_7927 View specimen

|
M3#1374Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1374 state:published |
Download 3D surface file |
Mus musculus Tourch_7932 View specimen

|
M3#1375Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1375 state:published |
Download 3D surface file |
Mus musculus Bal02 View specimen

|
M3#1320Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1320 state:published |
Download 3D surface file |
Mus musculus Bal04 View specimen

|
M3#1321Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1321 state:published |
Download 3D surface file |
Mus musculus Bal06 View specimen

|
M3#1322Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1322 state:published |
Download 3D surface file |
Mus musculus Bal08 View specimen

|
M3#1323Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1323 state:published |
Download 3D surface file |
Mus musculus Bal11 View specimen

|
M3#1324Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1324 state:published |
Download 3D surface file |
Mus musculus Bal12 View specimen

|
M3#1325Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1325 state:published |
Download 3D surface file |
Mus musculus Bal15 View specimen

|
M3#1326Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1326 state:published |
Download 3D surface file |
Mus musculus Bal16 View specimen

|
M3#1327Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1327 state:published |
Download 3D surface file |
Mus musculus Bal17 View specimen

|
M3#1328Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1328 state:published |
Download 3D surface file |
Mus musculus Bal18 View specimen

|
M3#1329Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1329 state:published |
Download 3D surface file |
Mus musculus Bal19 View specimen

|
M3#1330Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1330 state:published |
Download 3D surface file |
Mus musculus Bal20 View specimen

|
M3#1331Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1331 state:published |
Download 3D surface file |
Mus musculus Bal21 View specimen

|
M3#1332Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1332 state:published |
Download 3D surface file |
Mus musculus Bal22 View specimen

|
M3#1333Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1333 state:published |
Download 3D surface file |
Mus musculus Bal23 View specimen

|
M3#1334Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1334 state:published |
Download 3D surface file |
Mus musculus Bal24 View specimen

|
M3#1335Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1335 state:published |
Download 3D surface file |
Mus musculus Bal25 View specimen

|
M3#1336Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1336 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_035 View specimen

|
M3#1337Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1337 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_046 View specimen

|
M3#1338Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1338 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_054 View specimen

|
M3#1339Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1339 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_056 View specimen

|
M3#1340Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1340 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_082 View specimen

|
M3#1341Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1341 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_086 View specimen

|
M3#1342Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1342 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_092 View specimen

|
M3#1343Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1343 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_319 View specimen

|
M3#1344Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1344 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_325 View specimen

|
M3#1345Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1345 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_329 View specimen

|
M3#1346Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1346 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_330 View specimen

|
M3#1347Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1347 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_F2b View specimen

|
M3#1348Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1348 state:published |
Download 3D surface file |
Mus musculus Balan_LAB_BB3weeks View specimen

|
M3#1349Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1349 state:published |
Download 3D surface file |
Mus musculus SW0ter View specimen

|
M3#1376Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1376 state:published |
Download 3D surface file |
Mus musculus SW343 View specimen

|
M3#1377Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1377 state:published |
Download 3D surface file |
Mus musculus SW1 View specimen

|
M3#1378Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1378 state:published |
Download 3D surface file |
Mus musculus SW2 View specimen

|
M3#1379Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1379 state:published |
Download 3D surface file |
Mus musculus BAL_F1_30x17_27j View specimen

|
M3#1350Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1350 state:published |
Download 3D surface file |
Mus musculus BAL_F1_167_48j View specimen

|
M3#1351Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1351 state:published |
Download 3D surface file |
Mus musculus BAL_F1_188_32j View specimen

|
M3#1352Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1352 state:published |
Download 3D surface file |
Mus musculus BAL_F1_192_28j View specimen

|
M3#1353Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1353 state:published |
Download 3D surface file |
Mus musculus BAL_F1_194_46j View specimen

|
M3#1354Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1354 state:published |
Download 3D surface file |
Mus musculus BAL_F1_196_44j View specimen

|
M3#1355Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1355 state:published |
Download 3D surface file |
Mus musculus BAL_F2_40x56_24j View specimen

|
M3#1356Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1356 state:published |
Download 3D surface file |
Mus musculus BAL_F2_47x61_22j View specimen

|
M3#1357Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1357 state:published |
Download 3D surface file |
Mus musculus Gardouch_3419 View specimen

|
M3#1358Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1358 state:published |
Download 3D surface file |
Mus musculus Gardouch_3432 View specimen

|
M3#1359Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1359 state:published |
Download 3D surface file |
Mus musculus Gardouch_3437 View specimen

|
M3#1360Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1360 state:published |
Download 3D surface file |
Mus musculus Gardouch_3439 View specimen

|
M3#1361Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1361 state:published |
Download 3D surface file |
Mus musculus Gardouch_3450 View specimen

|
M3#1362Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1362 state:published |
Download 3D surface file |
Mus musculus Gardouch_3453 View specimen

|
M3#1363Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1363 state:published |
Download 3D surface file |
Mus musculus Gardouch_3459 View specimen

|
M3#1364Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1364 state:published |
Download 3D surface file |
Mus musculus Gardouch_3462 View specimen

|
M3#1365Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1365 state:published |
Download 3D surface file |
Mus musculus SW5 View specimen

|
M3#1380Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1380 state:published |
Download 3D surface file |
Mus musculus SWF3 View specimen

|
M3#1381Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1381 state:published |
Download 3D surface file |
Mus musculus SW342 View specimen

|
M3#1382Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1382 state:published |
Download 3D surface file |
Mus musculus SW341 View specimen

|
M3#1383Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1383 state:published |
Download 3D surface file |
Mus musculus SW339 View specimen

|
M3#1384Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1384 state:published |
Download 3D surface file |
Mus musculus SWF4 View specimen

|
M3#1385Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1385 state:published |
Download 3D surface file |
Mus musculus SW0bis_350 View specimen

|
M3#1386Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1386 state:published |
Download 3D surface file |
Mus musculus SW0_348 View specimen

|
M3#1387Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1387 state:published |
Download 3D surface file |
Mus musculus SW347 View specimen

|
M3#1388Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1388 state:published |
Download 3D surface file |
Mus musculus SW345 View specimen

|
M3#1389Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1389 state:published |
Download 3D surface file |
Mus musculus hyb_125xSW_01 View specimen

|
M3#1390Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1390 state:published |
Download 3D surface file |
Mus musculus hyb_125xSW_02 View specimen

|
M3#1391Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1391 state:published |
Download 3D surface file |
Mus musculus hyb_SWx126_01 View specimen

|
M3#1392Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1392 state:published |
Download 3D surface file |
Mus musculus hyb_SWx126_02 View specimen

|
M3#1393Two bony labyrinths Type: "3D_surfaces"doi: 10.18563/m3.sf.1393 state:published |
Download 3D surface file |

This contribution contains the 3D models described and figured in the following publication: Gaetano, L. C., Abdala, F., Mancuso, C, and Vega N.2025. New traversodontid cynodont from the Late Triassic Chañares Formation. Publicación Electrónica de la Asociación Paleontológica Argentina.
Pontognathus ignotus PULR-V 287 View specimen

|
M3#1647partial snout preserving the lateralmost incisor, the base of the canine, and several postcanines Type: "3D_surfaces"doi: 10.18563/m3.sf.1647 state:published |
Download 3D surface file |
Massetognathus pascuali PULR-V 289 View specimen

|
M3#1646partial lower jaw Type: "3D_surfaces"doi: 10.18563/m3.sf.1646 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models analyzed in the publication: Mummified Paleogene Spirostreptida and Julida (Arthropoda, Diplopoda) from southern France. Papers in Paleontology.
Protosilvestria sculpta NMB F1935 View specimen

|
M3#1457Paralectotype, 13 diplosegments with the proximal part of the legs Type: "3D_surfaces"doi: 10.18563/m3.sf.1457 state:published |
Download 3D surface file |

|
M3#1657CT data of NMB F1935. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1657 state:published |
Download CT data |
Protosilvestria sculpta NMB F1936 View specimen

|
M3#1458Lectotype, head with the ten following segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1458 state:published |
Download 3D surface file |

|
M3#1658CT data of NMB F1936. Type: "3D_CT"doi: 10.18563/m3.sf.1658 state:published |
Download CT data |
Protosilvestria sculpta NMB F1937 View specimen

|
M3#1459Paralectotype, seven segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1459 state:published |
Download 3D surface file |

|
M3#1659CT data of NMB F1937 Type: "3D_CT"doi: 10.18563/m3.sf.1659 state:published |
Download CT data |
Protosilvestria sculpta NMB F1938 View specimen

|
M3#1460Paralectotype, ten segments and the telson Type: "3D_surfaces"doi: 10.18563/m3.sf.1460 state:published |
Download 3D surface file |

|
M3#1660CT data of NMB F1938 Type: "3D_CT"doi: 10.18563/m3.sf.1660 state:published |
Download CT data |
Protosilvestria sculpta NMB F1987 View specimen

|
M3#1461Nine segments and the telson Type: "3D_surfaces"doi: 10.18563/m3.sf.1461 state:published |
Download 3D surface file |

|
M3#1661CT data of NMB F1987 Type: "3D_CT"doi: 10.18563/m3.sf.1661 state:published |
Download CT data |
Protosilvestria sculpta NMB F1988 View specimen

|
M3#1462Two parts, first part=telson and four segments, second part=five segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1462 state:published |
Download 3D surface file |

|
M3#1662CT data of NMB F1988 Type: "3D_CT"doi: 10.18563/m3.sf.1662 state:published |
Download CT data |
Protosilvestria sculpta NMB F1989 View specimen

|
M3#1463Telson with 13 segments and the digestive tract Type: "3D_surfaces"doi: 10.18563/m3.sf.1463 state:published |
Download 3D surface file |

|
M3#1663CT data of NMB F1989 Type: "3D_CT"doi: 10.18563/m3.sf.1663 state:published |
Download CT data |
Protosilvestria sculpta NMB F1990 View specimen

|
M3#1464Eight segments and the telson Type: "3D_surfaces"doi: 10.18563/m3.sf.1464 state:published |
Download 3D surface file |

|
M3#1664CT data of NMB F1990 Type: "3D_CT"doi: 10.18563/m3.sf.1664 state:published |
Download CT data |
Protosilvestria sculpta NMB F3743 View specimen

|
M3#1465Seven segments and legs Type: "3D_surfaces"doi: 10.18563/m3.sf.1465 state:published |
Download 3D surface file |

|
M3#1665CT data of NMB F3743. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1665 state:published |
Download CT data |
Protosilvestria sculpta UM-SND-1704 View specimen

|
M3#1468Head and seven segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1468 state:published |
Download 3D surface file |

|
M3#1666CT data of UM-SND-1704. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1666 state:published |
Download CT data |
Indet Indet UM-ROQ1-500 View specimen

|
M3#1467Head and ten segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1467 state:published |
Download 3D surface file |

|
M3#1667CT data of UM-ROQ1-500. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1667 state:published |
Download CT data |
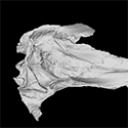
The present 3D Dataset contains the 3D model analyzed in The largest freshwater odontocete: a South Asian river dolphin relative from the Proto-Amazonia.
Pebanista yacuruna MUSM 4017 View specimen

|
M3#1394Holotype skull of Pebanista yacuruna MUSM 4017 Type: "3D_surfaces"doi: 10.18563/m3.sf.1394 state:published |
Download 3D surface file |

The holotype of Hamadasuchus rebouli Buffetaut 1994 from the Kem Kem beds of Morocco (Late Albian – Cenomanian) consists of a left dentary which is limited, fragmentary and reconstructed in some areas. To aid in assessing if the original diagnosis can be considered as valid, the specimen was CT scanned for the first time. This is especially important to resolve the taxonomic status of certain specimens that have been assigned to Hamadasuchus rebouli since then. The reconstructed structures in this contribution are in agreement with the original description, notably in terms of alveolar count; thus the original diagnosis of this taxon remains valid and some specimens are not referable to H. rebouli anymore.
Hamadasuchus rebouli MDE C001 View specimen

|
M3#1402Dentary and teeth Type: "3D_surfaces"doi: 10.18563/m3.sf.1402 state:published |
Download 3D surface file |

|
M3#1403Toothmarks Type: "3D_surfaces"doi: 10.18563/m3.sf.1403 state:published |
Download 3D surface file |
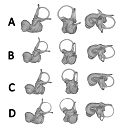
The present 3D Dataset contains the 3D models analyzed in the following publication: Size variation under domestication: Conservatism in the inner ear shape of wolves, dogs and dingoes. Scientific Reports 7, Article number: 13330, https://doi.org/10.1038/s41598-017-13523-9.
Canis lupus familiaris NMBE 16 View specimen

|
M3#2293D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.229 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE-LAT-1136 View specimen

|
M3#2423D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.242 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE-LAT-1119 View specimen

|
M3#2433D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.243 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE-BUR-1057 View specimen

|
M3#2443D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.244 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE-LUS-1102 View specimen

|
M3#2453D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.245 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE-LUS-1095 View specimen

|
M3#2463D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.246 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE-DUR-1124 View specimen

|
M3#2473D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.247 state:published |
Download 3D surface file |
Canis lupus chanco ZMUZH 17603 View specimen

|
M3#2483D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.248 state:published |
Download 3D surface file |
Canis lupus chanco ZMUZH 20201 View specimen

|
M3#2493D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.249 state:published |
Download 3D surface file |
Canis lupus chanco ZMUZH 17602 View specimen

|
M3#2503D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.250 state:published |
Download 3D surface file |
Canis lupus ZMUZH 13854 View specimen

|
M3#2403D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.240 state:published |
Download 3D surface file |
Canis lupus chanco ZMUZH 20202 View specimen

|
M3#2393D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.239 state:published |
Download 3D surface file |
Canis lupus chanco ZMUZH 17612 View specimen

|
M3#2303D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.230 state:published |
Download 3D surface file |
Canis lupus chanco ZMUZH 18082 View specimen

|
M3#2313D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.231 state:published |
Download 3D surface file |
Canis lupus ZMUZH 17118 View specimen

|
M3#2323D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.232 state:published |
Download 3D surface file |
Canis lupus ZMUZH 15858 View specimen

|
M3#2333D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.233 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 17712 View specimen

|
M3#2343D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.234 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 17713 View specimen

|
M3#2353D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.235 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 10166 View specimen

|
M3#2363D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.236 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 10175 View specimen

|
M3#2373D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.237 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 14842 View specimen

|
M3#2383D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.238 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 10342 View specimen

|
M3#2513D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.251 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 10343 View specimen

|
M3#2523D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.252 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 13766 View specimen

|
M3#2533D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.253 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 17717 View specimen

|
M3#2653D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.265 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 17711 View specimen

|
M3#2663D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.266 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 17714 View specimen

|
M3#2673D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.267 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH 17715 View specimen

|
M3#2683D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.268 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2835 View specimen

|
M3#2693D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.269 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2834 View specimen

|
M3#2703D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.270 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2837 View specimen

|
M3#2713D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.271 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2831 View specimen

|
M3#2723D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.272 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2845 View specimen

|
M3#2733D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.273 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 3001 View specimen

|
M3#2643D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.264 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2832 View specimen

|
M3#2633D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.263 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 3000 View specimen

|
M3#2543D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.254 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2847 View specimen

|
M3#2553D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.255 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2846 View specimen

|
M3#2563D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.256 state:published |
Download 3D surface file |
Canis lupus familiaris PIMUZ A/V 2836 View specimen

|
M3#2573D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.257 state:published |
Download 3D surface file |
Canis lupus familiaris NMB 12080 View specimen

|
M3#2583D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.258 state:published |
Download 3D surface file |
Canis lupus familiaris NMB 12081 View specimen

|
M3#2593D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.259 state:published |
Download 3D surface file |
Canis lupus familiaris NMB 12079 View specimen

|
M3#2603D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.260 state:published |
Download 3D surface file |
Canis lupus familiaris NMB 12078 View specimen

|
M3#2613D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.261 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE 1051209 View specimen

|
M3#2623D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.262 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE 1051226 View specimen

|
M3#2283D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.228 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE 1051381 View specimen

|
M3#2213D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.221 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE 1051418 View specimen

|
M3#1843D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.184 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH A.II. View specimen

|
M3#1973D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.197 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH A.VII. View specimen

|
M3#1983D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.198 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH We.6. View specimen

|
M3#1993D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.199 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH Ez.2. View specimen

|
M3#2003D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.200 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH Ez.E. View specimen

|
M3#2013D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.201 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH A.6. View specimen

|
M3#2023D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.202 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH Wyn.9. View specimen

|
M3#2033D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.203 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH F.48. View specimen

|
M3#2043D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.204 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH Terp.1. View specimen

|
M3#2053D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.205 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH A.VIII. View specimen

|
M3#1963D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.196 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH A.VI. View specimen

|
M3#1953D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.195 state:published |
Download 3D surface file |
Canis lupus familiaris ZMUZH A.IV. View specimen

|
M3#1853D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.185 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE A.403. View specimen

|
M3#1873D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.187 state:published |
Download 3D surface file |
Canis lupus familiaris NMBE A.5.a. View specimen

|
M3#1883D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.188 state:published |
Download 3D surface file |
Canis lupus NMB 8381 View specimen

|
M3#1893D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.189 state:published |
Download 3D surface file |
Canis lupus lycaon NMB C.1362 View specimen

|
M3#1903D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.190 state:published |
Download 3D surface file |
Canis lupus NMB Z309 View specimen

|
M3#1913D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.191 state:published |
Download 3D surface file |
Canis lupus NMB 2761 View specimen

|
M3#1923D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.192 state:published |
Download 3D surface file |
Canis lupus occidentalis NMB No Nb View specimen

|
M3#1933D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.193 state:published |
Download 3D surface file |
Canis lupus NMB 5258 View specimen

|
M3#1943D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.194 state:published |
Download 3D surface file |
Canis lupus NMB SCM320 View specimen

|
M3#2063D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.206 state:published |
Download 3D surface file |
Canis lupus arabs NMB 11019 View specimen

|
M3#2073D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.207 state:published |
Download 3D surface file |
Canis lupus UMZC K.3141 View specimen

|
M3#2083D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.208 state:published |
Download 3D surface file |
Canis lupus UMZC K.3150.1 View specimen

|
M3#2193D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.219 state:published |
Download 3D surface file |
Canis lupus UMZC K.3152 View specimen

|
M3#2203D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.220 state:published |
Download 3D surface file |
Canis lupus UMZC K.3149 View specimen

|
M3#2223D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.222 state:published |
Download 3D surface file |
Canis lupus familiaris UMZC K.3016 View specimen

|
M3#2233D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.223 state:published |
Download 3D surface file |
Canis lupus occidentalis ZMUZH 17210 View specimen

|
M3#2243D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.224 state:published |
Download 3D surface file |
Canis lupus familiaris SZ 7961 View specimen

|
M3#2253D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.225 state:published |
Download 3D surface file |
Canis lupus familiaris SZ 7959 View specimen

|
M3#2263D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.226 state:published |
Download 3D surface file |
Canis lupus familiaris SZ 7958 View specimen

|
M3#2173D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.217 state:published |
Download 3D surface file |
Canis lupus familiaris SZ 7930 View specimen

|
M3#2163D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.216 state:published |
Download 3D surface file |
Canis lupus familiaris SZ 7926 View specimen

|
M3#2183D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.218 state:published |
Download 3D surface file |
Canis lupus familiaris SZ 7929 View specimen

|
M3#2093D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.209 state:published |
Download 3D surface file |
Canis lupus dingo M6297 View specimen

|
M3#1863D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.186 state:published |
Download 3D surface file |
Canis lupus dingo M24153 View specimen

|
M3#2103D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.210 state:published |
Download 3D surface file |
Canis lupus dingo M33608 View specimen

|
M3#2113D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.211 state:published |
Download 3D surface file |
Canis lupus dingo M38587 View specimen

|
M3#2123D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.212 state:published |
Download 3D surface file |
Canis lupus dingo Blumenbach UMZC K.3221 View specimen

|
M3#2133D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.213 state:published |
Download 3D surface file |
Canis lupus dingo Blumenbach UMZC K.3223 View specimen

|
M3#2143D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.214 state:published |
Download 3D surface file |
Canis lupus dingo UniSyd FVS 45 View specimen

|
M3#2153D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.215 state:published |
Download 3D surface file |
Canis lupus dingo UNSW Z354 View specimen

|
M3#2273D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.227 state:published |
Download 3D surface file |
Canis lupus familiaris TMM M-150 View specimen

|
M3#2413D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.241 state:published |
Download 3D surface file |
Canis lupus M39960 View specimen

|
M3#2743D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.274 state:published |
Download 3D surface file |
Canis lupus NMB 8635 View specimen

|
M3#2753D virtual endocast of the left inner ear Type: "3D_surfaces"doi: 10.18563/m3.sf.275 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models analyzed in Assemat et al. 2023: Shape diversity in conodont elements, a quantitative study using 3D topography. Marine Micropaleontology 184. https://doi.org/10.1016/j.marmicro.2023.102292
P1 elements represent dental components of the conodont apparatus that perform the final stage of food processing before ingestion. Consequently, quantifying the shape of P1 elements across the topographic indices of different conodont species becomes crucial for deciphering the diversity in feeding behavior within this group.
Bispathodus aculeatus UM CTB 082 View specimen

|
M3#1404P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1404 state:published |
Download 3D surface file |
Bispathodus aculeatus UM CTB 083 View specimen

|
M3#1405P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1405 state:published |
Download 3D surface file |
Bispathodus aculeatus UM CTB 086 View specimen

|
M3#1406P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1406 state:published |
Download 3D surface file |
Bispathodus ultimus UM CTB 088 View specimen

|
M3#1407P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1407 state:published |
Download 3D surface file |
Bispathodus aculeatus UM CTB 089 View specimen

|
M3#1408P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1408 state:published |
Download 3D surface file |
Bispathodus costatus UM CTB 090 View specimen

|
M3#1409P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1409 state:published |
Download 3D surface file |
Bispathodus ultimus UM CTB 092 View specimen

|
M3#1410P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1410 state:published |
Download 3D surface file |
Bispathodus costatus UM CTB 093 View specimen

|
M3#1411P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1411 state:published |
Download 3D surface file |
Bispathodus spinulicostatus UM CTB 094 View specimen

|
M3#1412P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1412 state:published |
Download 3D surface file |
Bispathodus aculeatus UM CTB 096 View specimen

|
M3#1413P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1413 state:published |
Download 3D surface file |
Bispathodus ultimus UM CTB 098 View specimen

|
M3#1414P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1414 state:published |
Download 3D surface file |
Bispathodus costatus UM CTB 060 View specimen

|
M3#1415P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1415 state:published |
Download 3D surface file |
Bispathodus spinulicostatus UM CTB 073 View specimen

|
M3#1416P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1416 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 049 View specimen

|
M3#1417P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1417 state:published |
Download 3D surface file |
Branmehla inornata UM CTB 100 View specimen

|
M3#1418P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1418 state:published |
Download 3D surface file |
Bispathodus stabilis (morphe 1) UM CTB 101 View specimen

|
M3#1419P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1419 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 102 View specimen

|
M3#1420P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1420 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 103 View specimen

|
M3#1421P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1421 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 104 View specimen

|
M3#1422P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1422 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 105 View specimen

|
M3#1423P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1423 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 106 View specimen

|
M3#1424P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1424 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 072 View specimen

|
M3#1425P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1425 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 107 View specimen

|
M3#1426P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1426 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 108 View specimen

|
M3#1427P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1427 state:published |
Download 3D surface file |
Branmehla suprema UM CTB 109 View specimen

|
M3#1428P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1428 state:published |
Download 3D surface file |
Bispathodus stabilis (morphe 1) UM CTB 110 View specimen

|
M3#1429P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1429 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 112 View specimen

|
M3#1430P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1430 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 061 View specimen

|
M3#1431P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1431 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 115 View specimen

|
M3#1432P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1432 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 116 View specimen

|
M3#1433P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1433 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 117 View specimen

|
M3#1434P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1434 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 062 View specimen

|
M3#1435P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1435 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 118 View specimen

|
M3#1436P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1436 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 119 View specimen

|
M3#1437P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1437 state:published |
Download 3D surface file |
Palmatolepis gracilis UM CTB 120 View specimen

|
M3#1438P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1438 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 075 View specimen

|
M3#1439P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1439 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 121 View specimen

|
M3#1440P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1440 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 122 View specimen

|
M3#1441P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1441 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 123 View specimen

|
M3#1442P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1442 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 125 View specimen

|
M3#1443P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1443 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 126 View specimen

|
M3#1444P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1444 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 128 View specimen

|
M3#1445P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1445 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 130 View specimen

|
M3#1446P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1446 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 131 View specimen

|
M3#1447P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1447 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 132 View specimen

|
M3#1448P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1448 state:published |
Download 3D surface file |
Polygnathus communis UM CTB 133 View specimen

|
M3#1449P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1449 state:published |
Download 3D surface file |
Polygnathus symmetricus UM CTB 139 View specimen

|
M3#1450P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1450 state:published |
Download 3D surface file |
Polygnathus symmetricus UM CTB 140 View specimen

|
M3#1451P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1451 state:published |
Download 3D surface file |
Polygnathus symmetricus UM CTB 141 View specimen

|
M3#1452P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1452 state:published |
Download 3D surface file |
Polygnathus symmetricus UM CTB 142 View specimen

|
M3#1453P element Type: "3D_surfaces"doi: 10.18563/m3.sf.1453 state:published |
Download 3D surface file |
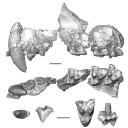
This contribution contains the three-dimensional models of the most complete and/or informative fossil materials attributed to Peradectes crocheti Gernelle, 2024, the earliest peradectid metatherian species of Europe, from its type locality (Palette, Provence, ~55 Ma). These specimens were analyzed and discussed in: Gernelle et al. (2024), Taxonomy and evolutionary history of peradectids (Metatheria): new data from the early Eocene of France. https://doi.org/10.1007/s10914-024-09724-5
Peradectes crocheti MHN.AIX.PV.2018.26.14 View specimen

|
M3#14993D surface model of MHN.AIX.PV.2018.26.14, fragmentary left maxilla with C-P1, anterior root of P2, and M1-M3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1499 state:published |
Download 3D surface file |
Peradectes crocheti MHN.AIX.PV.2017.6.6 View specimen

|
M3#15003D surface model of MHN.AIX.PV.2017.6.6, left P2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1500 state:published |
Download 3D surface file |
Peradectes crocheti MHN.AIX.PV.2017.6.7 View specimen

|
M3#15013D surface model of MHN.AIX.PV.2017.6.7, left M3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1501 state:published |
Download 3D surface file |
Peradectes crocheti MHN.AIX.PV.2017.6.8 View specimen

|
M3#15023D surface model of MHN.AIX.PV.2017.6.8, right hemi-mandible fragment with canine alveolus, posterior root of p1, partial p2, p3, partial m1, and m2-m3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1502 state:published |
Download 3D surface file |
Peradectes crocheti MHN.AIX.PV.2017.6.9 View specimen

|
M3#15033D surface model of MHN.AIX.PV.2017.6.9, leftm1-m4 row with fragments of dentary Type: "3D_surfaces"doi: 10.18563/m3.sf.1503 state:published |
Download 3D surface file |
Peradectes crocheti MHN.AIX.PV.2017.6.14 View specimen

|
M3#15043D surface model of MHN.AIX.PV.2017.6.14, right astragalus Type: "3D_surfaces"doi: 10.18563/m3.sf.1504 state:published |
Download 3D surface file |
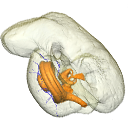
In this contribution, we describe the external and internal morphology of a delphinid petrosal bone collected from Ahu Tahai, a burial site located on the Southwestern coast of Easter Island, at Hangaroa. We discuss the taxonomic attribution of this archaeological item and describe its internal structures based on µCT data, including the bony labyrinth and the nerve and vein patterns. Identification of the nerves exists lead us to relocate the identification of the foramen singulare in delphinid petrosals.
indet. indet. AT1 View specimen

|
M3#420Stapes Type: "3D_surfaces"doi: 10.18563/m3.sf.420 state:published |
Download 3D surface file |
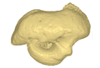
|
M3#421petrosal bone Type: "3D_surfaces"doi: 10.18563/m3.sf.421 state:published |
Download 3D surface file |
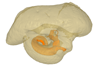
|
M3#422in situ bony labyrinth Type: "3D_surfaces"doi: 10.18563/m3.sf.422 state:published |
Download 3D surface file |
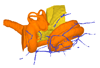
|
M3#423bony labyrinth and associated nerves and blood vessels Type: "3D_surfaces"doi: 10.18563/m3.sf.423 state:published |
Download 3D surface file |

This contribution contains the three-dimensional digital model of one isolated fossil tooth of an anthropoid primate (Ashaninkacebus simpsoni), discovered in sedimentary deposits located on the upper Rio Juruá in State of Acre, Brazil (Western Amazonia). This fossil was described, figured and discussed in the following publication: Marivaux et al. (2023), An eosimiid primate of South Asian affinities in the Paleogene of Western Amazonia and the origin of New World monkeys. Proceedings of the National Academy of Sciences USA. https://doi.org/10.1073/pnas.2301338120
Ashaninkacebus simpsoni UFAC-CS 066 View specimen
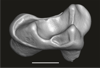
|
M3#1114Right first upper molar (rM1), pristine. Type: "3D_surfaces"doi: 10.18563/m3.sf.1114 state:published |
Download 3D surface file |
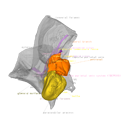
This project presents the osteological connexions of the petrosal bone of the extant Hippopotamidae Hippopotamus amphibius and Choeropsis liberiensis by a virtual osteological dissection of the ear region. The petrosal, the bulla, the sinuses and the major morphological features surrounding the petrosal bone are labelled, both in situ and in an exploded model presenting disassembly views. The directional underwater hearing mode of Hippopotamidae is discussed based on the new observations.
Choeropsis liberiensis UPPal-M09-5-005a View specimen
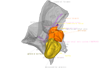
|
M3#1Labelled compact model of the right ear region of Choeropsis liberiensis (UPPal-M09-5-005a) Type: "3D_surfaces"doi: 10.18563/m3.sf1 state:published |
Download 3D surface file |

|
M3#2Labelled exploded model of the right ear region of Choeropsis liberiensis (UPPal-M09-5-005a) Type: "3D_surfaces"doi: 10.18563/m3.sf2 state:published |
Download 3D surface file |
Hippopotamus amphibius UM N179 View specimen
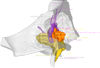
|
M3#3Labelled compact model of the right ear region of Hippopotamus amphibius (UM N 179) Type: "3D_surfaces"doi: 10.18563/m3.sf3 state:published |
Download 3D surface file |

|
M3#4Labelled exploded model of the right ear region of Hippopotamus amphibius (UM N 179) Type: "3D_surfaces"doi: 10.18563/m3.sf4 state:published |
Download 3D surface file |
This contribution contains the three-dimensional models of the inner ear of the hetaxodontid rodents Amblyrhiza, Clidomys and Elasmodontomys from the West Indies. These specimens were analyzed and discussed in : The inner ear of caviomorph rodents: phylogenetic implications and application to extinct West Indian taxa.
Amblyrhiza inundata 11842 View specimen

|
M3#11543D surface of the left-oriented inner ear of Amblyrhiza. Type: "3D_surfaces"doi: 10.18563/m3.sf.1154 state:published |
Download 3D surface file |
Clidomys sp NA View specimen
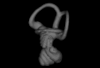
|
M3#11553D surface of the left-oriented inner ear of Clidomys sp. Type: "3D_surfaces"doi: 10.18563/m3.sf.1155 state:published |
Download 3D surface file |
Elasmodontomys obliquus 17127 View specimen
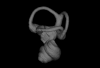
|
M3#11563D surface of the left-oriented inner ear of Elasmodontomys obliquus. Type: "3D_surfaces"doi: 10.18563/m3.sf.1156 state:published |
Download 3D surface file |

This contribution contains 3D models of the holotype of a new species of long-nosed armadillos, the Guianan long-nosed armadillo (Dasypus guianensis) described in the following publication: Barthe M., Rancilhac L., Arteaga M. C., Feijó A., Tilak M.-K., Justy F., Loughry W. J., McDonough C. M., de Thoisy B., Catzeflis F., Billet G., Hautier L., Nabholz B., and Delsuc F. 2024. Exon capture museomics deciphers the nine-banded armadillo species complex and identifies a new species endemic to the Guiana Shield. Systematic Biology, syae027. https://doi.org/10.1093/sysbio/syae027
Dasypus guianensis MNHN-ZM-MO-2001-1317 View specimen

|
M3#1200Skeleton and carapace Type: "3D_surfaces"doi: 10.18563/m3.sf.1200 state:published |
Download 3D surface file |

|
M3#1201Frontal sinuses Type: "3D_surfaces"doi: 10.18563/m3.sf.1201 state:published |
Download 3D surface file |
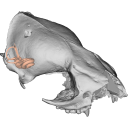
The present 3D Dataset contains 3D models of the cranium surface and of the bony labyrinth endocast of the stem bat Vielasia sigei. They are used by (Hand et al., 2023) to explore the phylogenetic position of this species, to infer its laryngeal echolocating capabilities, and to eventually discuss chiropteran evolution before the crown clade diversification.
Vielasia sigei UM VIE-250 View specimen

|
M3#1269External surface of the cranium Type: "3D_surfaces"doi: 10.18563/m3.sf.1269 state:published |
Download 3D surface file |
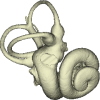
|
M3#1270Virtual endocast of the right bony labyrinth Type: "3D_surfaces"doi: 10.18563/m3.sf.1270 state:published |
Download 3D surface file |