Explodable 3D Dog Skull for Veterinary Education
3D models of a Sheep and Goat Skull and Inner ear
3D models of Miocene vertebrates from Tavers
3D GM dataset of bird skeletal variation
Skeletal embryonic development in the catshark
Bony connexions of the petrosal bone of extant hippos
bony labyrinth (11) , inner ear (10) , Eocene (8) , South America (8) , Paleobiogeography (7) , skull (7) , phylogeny (6)
Lionel Hautier (23) , Maëva Judith Orliac (21) , Laurent Marivaux (16) , Rodolphe Tabuce (14) , Bastien Mennecart (13) , Pierre-Olivier Antoine (12) , Renaud Lebrun (11)
|
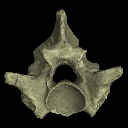
3D data related to the publication: A new species of Palaeopython (Serpentes) and other extinct squamates from the Eocene of Dielsdorf (Zurich, Switzerland)
|

|
M3#399ZIP file containing .ply of vertebra PIMUZ A/III 631 from Palaeopython helveticus n. sp. Type: "3D_surfaces"doi: 10.18563/m3.sf.399 state:published |
Download 3D surface file |

|
M3#403dataset of snake vertebra PIMUZ A/III 631 Type: "3D_CT"doi: 10.18563/m3.sf.403 state:published |
Download CT data |
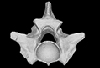
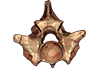
Palaeopython helveticus PIMUZ A/III 634 View specimen
|
M3#400ZIP file containing .ply of vertebra PIMUZ A/III 634 from Palaeopython helveticus n. sp. (holotype) Type: "3D_surfaces"doi: 10.18563/m3.sf.400 state:published |
Download 3D surface file |
|
M3#404dataset of snake vertbra PIMUZ A/III 634 (holotype) Type: "3D_CT"doi: 10.18563/m3.sf.404 state:published |
Download CT data |
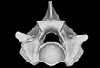
Palaeopython helveticus PIMUZ A/III 636 View specimen
|
M3#401ZIP file containing .ply of vertebra PIMUZ A/III 636 from Palaeopython helveticus n. sp. Type: "3D_surfaces"doi: 10.18563/m3.sf.401 state:published |
Download 3D surface file |
|
M3#406dataset of snake vertebra PIMUZ A/III 636 Type: "3D_CT"doi: 10.18563/m3.sf.406 state:published |
Download CT data |
Palaeovaranus sp. PIMUZ A/III 234 View specimen

|
M3#402ZIP file containing .ply of dentary PIMUZ A/III 234 of Palaeovaranus sp. Type: "3D_surfaces"doi: 10.18563/m3.sf.402 state:published |
Download 3D surface file |

|
M3#405dataset of dentary of Palaeovaranus sp. (PIMUZ A/III 234) Type: "3D_CT"doi: 10.18563/m3.sf.405 state:published |
Download CT data |
This contribution contains 3D models of extinct rodents Dinomyidae from Miocene and Quaternary of Brazil. The Miocene specimens that were digitalized include the holotypes of Potamarchus adamiae, Pseudopotamarchus villanuevai, and Ferigolomys pacarana collected in the Solimões Formation (Upper Miocene), northern Brazil. The Quaternary specimens are the holotype and paratype of Niedemys piauiensis, found in Upper Pleistocene deposits from northeast Brazil.
Potamarchus adamiae UFAC-CS 011 View specimen

|
M3#410UFAC-CS 011 – holotype, palatal region of the skull with cheek teeth Type: "3D_surfaces"doi: 10.18563/m3.sf.410 state:published |
Download 3D surface file |
Potamarchus adamiae UFAC-CS 043 View specimen
|
M3#411UFAC-CS 043, left dentary with cheek teeth Type: "3D_surfaces"doi: 10.18563/m3.sf.411 state:published |
Download 3D surface file |
Pseudopotamarchus villanuevai UFAC 4762 View specimen

|
M3#412UFAC 4762 – holotype, incomplete right maxilla with cheek teeth Type: "3D_surfaces"doi: 10.18563/m3.sf.412 state:published |
Download 3D surface file |
Ferigolomys pacarana UFAC 6460 View specimen

|
M3#413UFAC 6460 – holotype, palatal region of the skull with cheek teeth Type: "3D_surfaces"doi: 10.18563/m3.sf.413 state:published |
Download 3D surface file |
Drytomomys sp. UFAC 2742 View specimen

|
M3#414UFAC 2742, right dentary with cheek teeth Type: "3D_surfaces"doi: 10.18563/m3.sf.414 state:published |
Download 3D surface file |
Niedemys piauiensis FUMDHAM 113-146365-2 View specimen

|
M3#418FUMDHAM 113-146365-2 - holotype, upper right tooth Type: "3D_surfaces"doi: 10.18563/m3.sf.418 state:published |
Download 3D surface file |
Niedemys piauiensis FUMDHAM 113-145304-2 View specimen
|
M3#419FUMDHAM 113-145304-2 - paratype, left lower molar Type: "3D_surfaces"doi: 10.18563/m3.sf.419 state:published |
Download 3D surface file |

This contribution contains the 3D models of the fossil teeth of two chinchilloid caviomorph rodents (Borikenomys praecursor and Chinchilloidea gen. et sp. indet.) discovered from lower Oligocene deposits of Puerto Rico, San Sebastian Formation (locality LACM Loc. 8060). These fossils were described and figured in the following publication: Marivaux et al. (2020), Early Oligocene chinchilloid caviomorphs from Puerto Rico and the initial rodent colonization of the West Indies. Proceedings of the Royal Society B. http://dx.doi.org/10.1098/rspb.2019.2806
Borikenomys praecursor LACM 162447 View specimen

|
M3#638Right lower m3. This isolated tooth was scanned with a resolution of 6 µm using a μ-CT-scanning station EasyTom 150 / Rx Solutions (Montpellier RIO Imaging, ISE-M, Montpellier, France). AVIZO 7.1 (Visualization Sciences Group) software was used for visualization, segmentation, and 3D rendering. The specimen was prepared within a “labelfield” module of AVIZO, using the segmentation threshold selection tool. Type: "3D_surfaces"doi: 10.18563/m3.sf.638 state:published |
Download 3D surface file |
Borikenomys praecursor LACM 162446 View specimen

|
M3#639Fragment of lower molar (most of the mesial part). This isolated broken tooth was scanned with a resolution of 6 µm using a μ-CT-scanning station EasyTom 150 / Rx Solutions (Montpellier RIO Imaging, ISE-M, Montpellier, France). AVIZO 7.1 (Visualization Sciences Group) software was used for visualization, segmentation, and 3D rendering. The specimen was prepared within a “labelfield” module of AVIZO, using the segmentation threshold selection tool. Type: "3D_surfaces"doi: 10.18563/m3.sf.639 state:published |
Download 3D surface file |
indet indet LACM 162448 View specimen

|
M3#640Fragment of either an upper tooth (mesial laminae) or a lower tooth (distal laminae). The specimen was scanned with a resolution of 6 µm using a μ-CT-scanning station EasyTom 150 / Rx Solutions (Montpellier RIO Imaging, ISE-M, Montpellier, France). AVIZO 7.1 (Visualization Sciences Group) software was used for visualization, segmentation, and 3D rendering. This fragment of tooth was prepared within a “labelfield” module of AVIZO, using the segmentation threshold selection tool. Type: "3D_surfaces"doi: 10.18563/m3.sf.640 state:published |
Download 3D surface file |

This contribution contains the 3D models of postcranial bones (humerus, ulna, innominate, femur, tibia, astragalus, navicular, and metatarsal III) described and figured in the following publication: “Postcranial morphology of the extinct rodent Neoepiblema (Rodentia: Chinchilloidea): insights into the paleobiology of neoepiblemids”.
Neoepiblema acreensis UFAC 3549 View specimen

|
M3#719UFAC 3549, left humerus missing the proximal region. Type: "3D_surfaces"doi: 10.18563/m3.sf.719 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 5076 View specimen

|
M3#720UFAC 5076, right humerus missing the proximal region. Type: "3D_surfaces"doi: 10.18563/m3.sf.720 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 1939 View specimen

|
M3#721UFAC 1939, right ulna missing the olecranon epiphysis and the distal region. Type: "3D_surfaces"doi: 10.18563/m3.sf.721 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 3697 View specimen

|
M3#722UFAC 3697, right innominate bone. Type: "3D_surfaces"doi: 10.18563/m3.sf.722 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 2574 View specimen
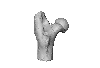
|
M3#723UFAC 2574, proximal region of a left femur. Type: "3D_surfaces"doi: 10.18563/m3.sf.723 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 2937 View specimen

|
M3#724UFAC 2937, right femur with damaged proximal region. Type: "3D_surfaces"doi: 10.18563/m3.sf.724 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 2210 View specimen

|
M3#725UFAC 2210, distal region of a right femur. Type: "3D_surfaces"doi: 10.18563/m3.sf.725 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 1887 View specimen

|
M3#726UFAC 1887, right tibia Type: "3D_surfaces"doi: 10.18563/m3.sf.726 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 1840 View specimen
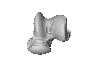
|
M3#727UFAC 1840, left astragalus. Type: "3D_surfaces"doi: 10.18563/m3.sf.727 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 2549 View specimen

|
M3#728UFAC 2549, right astragalus. Type: "3D_surfaces"doi: 10.18563/m3.sf.728 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 3672 View specimen

|
M3#729UFAC 3672, right navicular. Type: "3D_surfaces"doi: 10.18563/m3.sf.729 state:published |
Download 3D surface file |
Neoepiblema acreensis UFAC 2116 View specimen

|
M3#730UFAC 2116, left metatarsal III. Type: "3D_surfaces"doi: 10.18563/m3.sf.730 state:published |
Download 3D surface file |
Neoepiblema horridula UFAC 3260 View specimen

|
M3#731UFAC 3260, fragmented left innominate. Type: "3D_surfaces"doi: 10.18563/m3.sf.731 state:published |
Download 3D surface file |
Neoepiblema horridula UFAC 2620 View specimen

|
M3#732UFAC 2620, distal region of a right femur. Type: "3D_surfaces"doi: 10.18563/m3.sf.732 state:published |
Download 3D surface file |
Neoepiblema horridula UFAC 2737 View specimen

|
M3#733UFAC 2737, proximal region of right femur. Type: "3D_surfaces"doi: 10.18563/m3.sf.733 state:published |
Download 3D surface file |
Neoepiblema horridula UFAC 3202 View specimen

|
M3#734UFAC 3202, right tibia, missing the proximalmost and distal portions. Type: "3D_surfaces"doi: 10.18563/m3.sf.734 state:published |
Download 3D surface file |
Neoepiblema horridula UFAC 3212 View specimen

|
M3#735UFAC 3212, left astragalus. Type: "3D_surfaces"doi: 10.18563/m3.sf.735 state:published |
Download 3D surface file |

This contribution contains the 3D models described and figured in the following publication: Aguirre-Fernández G, Jost J, and Hilfiker S. 2022. First records of extinct kentriodontid and squalodelphinid dolphins from the Upper Marine Molasse (Burdigalian age) of Switzerland and a reappraisal of the Swiss cetacean fauna.
Kentriodon sp. NMBE 5023944 View specimen

|
M3#8583D models of left periotic and bony labyrinth of NMBE 5023944 (Kentriodon sp.) Type: "3D_surfaces"doi: 10.18563/m3.sf.858 state:published |
Download 3D surface file |
Kentriodon sp. NMBE 5023945 View specimen

|
M3#8593D models of right periotic and bony labyrinth of NMBE 5023945 (Kentriodontidae indet.) Type: "3D_surfaces"doi: 10.18563/m3.sf.859 state:published |
Download 3D surface file |
Kentriodon sp. NMBE 5023946 View specimen

|
M3#8603D models of left periotic and bony labyrinth of NMBE 5023946 (Kentriodon sp.) Type: "3D_surfaces"doi: 10.18563/m3.sf.860 state:published |
Download 3D surface file |
Kentriodon sp. NMBE 5036436 View specimen

|
M3#8613D models of right periotic and bony labyrinth of NMBE 5036436 (Kentriodontidae indet.) Type: "3D_surfaces"doi: 10.18563/m3.sf.861 state:published |
Download 3D surface file |
indet. indet. NMBE 5023942 View specimen

|
M3#8623D models of right periotic and bony labyrinth of NMBE 5023942 (Squalodelphinidae indet.) Type: "3D_surfaces"doi: 10.18563/m3.sf.862 state:published |
Download 3D surface file |
indet. indet. NMBE 5023943 View specimen

|
M3#8633D models of left periotic and bony labyrinth of NMBE 5023943 (Squalodelphinidae indet.) Type: "3D_surfaces"doi: 10.18563/m3.sf.863 state:published |
Download 3D surface file |
indet. indet. NMBE 5036437 View specimen

|
M3#8643D models of left periotic and bony labyrinth of NMBE 5036437 (Physeteridae indet.) Type: "3D_surfaces"doi: 10.18563/m3.sf.864 state:published |
Download 3D surface file |

The present 3D Dataset contains 3D models of the cranial, visceral, and pectoral endoskeleton of Iniopera, an iniopterygian stem-group holocephalan from the Pennsylvanian of the USA. These data formed the basis for the analyses carried out in Dearden et al. (2023) “Evidence for high-performance suction feeding in the Pennsylvanian stem-group holocephalan Iniopera” PNAS.
Iniopera sp. KUNHM 22060, 158289 View specimen

|
M3#1034plys of the head endoskeleton of Iniopera sp. Type: "3D_surfaces"doi: 10.18563/m3.sf.1034 state:published |
Download 3D surface file |
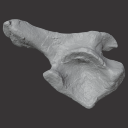
The present 3D Dataset contains the 3D models of an ilium, a vertebra, and a partial scapula of Prestosuchus sp. that were analyzed in “New Loricata remains from the Pinheiros-Chiniquá Sequence (Middle-Upper Triassic), southern Brazil”.
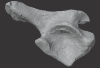
Prestosuchus sp. UFSM11603 View specimen
|
M3#1080Surface scan of a right ilium of Prestosuchus sp. with a 0.4 mm resolution. Type: "3D_surfaces"doi: 10.18563/m3.sf.1080 state:published |
Download 3D surface file |
Prestosuchus sp. UFSM11233 View specimen

|
M3#1081Surface scan of a partial right scapula of Prestosuchus sp. with a 0.4mm resolution. Type: "3D_surfaces"doi: 10.18563/m3.sf.1081 state:published |
Download 3D surface file |
Prestosuchus sp. UFSM11602a View specimen
|
M3#1082Surface scan of a anterior dorsal vertebra of Prestosuchus sp. with a 0.2 mm resolution. Type: "3D_surfaces"doi: 10.18563/m3.sf.1082 state:published |
Download 3D surface file |
The present 3D Dataset contains the 3D model analyzed in the publication : On Roth’s “human fossil” from Baradero, Buenos Aires Province, Argentina: morphological and genetic analysis. The “human fossil” from Baradero, Buenos Aires Province, Argentina, is a collection of skeleton parts first recovered by Swiss paleontologist Santiago Roth and further studied by anthropologist Rudolf Martin. By the end of the 19th century and beginning of the 20th century it was considered as one of the oldest human skeletons from the southern cone. We studied the cranial anatomy and contextualized the ancient individual remains. We discuss the context of the finding, conducted an osteobiographical assessment and performed a 3D virtual reconstruction of the skull, using micro-CT-scans on selected skull fragments and the mandible. This was followed by the extraction of bone tissue and teeth samples for radiocarbon and genetic analyses, which brought only limited results due to poor preservation and possible contamination. We estimate that the individual from Baradero is a middle-aged adult male. We conclude that the revision of foundational collections with current methodological tools brings new insights and clarifies long held assumptions on the significance of samples that were recovered when archaeology was not yet professionalized.
Homo sapiens PIMUZ A/V 4217 View specimen

|
M3#11983D virtual reconstruction of the skull Type: "3D_surfaces"doi: 10.18563/m3.sf.1198 state:published |
Download 3D surface file |

This contribution contains the 3D models described and figured in the following publication: Bonis et al. 2023. A new large pantherine and a sabre-toothed cat (Mammalia, Carnivora, Felidae) from the late Miocene hominoid-bearing Khorat sand pits, Nakhon Ratchasima Province, northeastern Thailand. The Science of Nature 110(5):42. https://doi.org/10.1007/s00114-023-01867-4
Pachypanthera piriyai CUF-KR-1 View specimen

|
M3#1209Holotype of Pachypanthera piriyai, a left hemi-mandible with alveoli for i1-i3 and canine, roots of p3, p4 and partially broken off m1 crown. Type: "3D_surfaces"doi: 10.18563/m3.sf.1209 state:published |
Download 3D surface file |
Pachypanthera piriyai CUF-KR-2 View specimen

|
M3#1210Paratype of Pachypanthera piriyai, a right hemi-maxilla with P3-P4, alveoli of C and M1, root of P2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1210 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models analyzed in the publication: Mummified Paleogene Spirostreptida and Julida (Arthropoda, Diplopoda) from southern France. Papers in Paleontology.
Protosilvestria sculpta NMB F1935 View specimen

|
M3#1457Paralectotype, 13 diplosegments with the proximal part of the legs Type: "3D_surfaces"doi: 10.18563/m3.sf.1457 state:published |
Download 3D surface file |

|
M3#1657CT data of NMB F1935. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1657 state:published |
Download CT data |
Protosilvestria sculpta NMB F1936 View specimen

|
M3#1458Lectotype, head with the ten following segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1458 state:published |
Download 3D surface file |

|
M3#1658CT data of NMB F1936. Type: "3D_CT"doi: 10.18563/m3.sf.1658 state:published |
Download CT data |
Protosilvestria sculpta NMB F1937 View specimen

|
M3#1459Paralectotype, seven segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1459 state:published |
Download 3D surface file |

|
M3#1659CT data of NMB F1937 Type: "3D_CT"doi: 10.18563/m3.sf.1659 state:published |
Download CT data |
Protosilvestria sculpta NMB F1938 View specimen

|
M3#1460Paralectotype, ten segments and the telson Type: "3D_surfaces"doi: 10.18563/m3.sf.1460 state:published |
Download 3D surface file |

|
M3#1660CT data of NMB F1938 Type: "3D_CT"doi: 10.18563/m3.sf.1660 state:published |
Download CT data |
Protosilvestria sculpta NMB F1987 View specimen

|
M3#1461Nine segments and the telson Type: "3D_surfaces"doi: 10.18563/m3.sf.1461 state:published |
Download 3D surface file |

|
M3#1661CT data of NMB F1987 Type: "3D_CT"doi: 10.18563/m3.sf.1661 state:published |
Download CT data |
Protosilvestria sculpta NMB F1988 View specimen

|
M3#1462Two parts, first part=telson and four segments, second part=five segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1462 state:published |
Download 3D surface file |

|
M3#1662CT data of NMB F1988 Type: "3D_CT"doi: 10.18563/m3.sf.1662 state:published |
Download CT data |
Protosilvestria sculpta NMB F1989 View specimen

|
M3#1463Telson with 13 segments and the digestive tract Type: "3D_surfaces"doi: 10.18563/m3.sf.1463 state:published |
Download 3D surface file |

|
M3#1663CT data of NMB F1989 Type: "3D_CT"doi: 10.18563/m3.sf.1663 state:published |
Download CT data |
Protosilvestria sculpta NMB F1990 View specimen

|
M3#1464Eight segments and the telson Type: "3D_surfaces"doi: 10.18563/m3.sf.1464 state:published |
Download 3D surface file |

|
M3#1664CT data of NMB F1990 Type: "3D_CT"doi: 10.18563/m3.sf.1664 state:published |
Download CT data |
Protosilvestria sculpta NMB F3743 View specimen

|
M3#1465Seven segments and legs Type: "3D_surfaces"doi: 10.18563/m3.sf.1465 state:published |
Download 3D surface file |

|
M3#1665CT data of NMB F3743. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1665 state:published |
Download CT data |
Protosilvestria sculpta UM-SND-1704 View specimen

|
M3#1468Head and seven segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1468 state:published |
Download 3D surface file |

|
M3#1666CT data of UM-SND-1704. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1666 state:published |
Download CT data |
Indet Indet UM-ROQ1-500 View specimen

|
M3#1467Head and ten segments Type: "3D_surfaces"doi: 10.18563/m3.sf.1467 state:published |
Download 3D surface file |

|
M3#1667CT data of UM-ROQ1-500. Images were reduced by a binning of factor 2. Type: "3D_CT"doi: 10.18563/m3.sf.1667 state:published |
Download CT data |

This contribution contains 3D models of the cranial endoskeleton of three specimens of the Permian ‘acanthodian’ stem-group chondrichthyan (cartilaginous fish) Acanthodes confusus, obtained using computed tomography. These datasets were described and analyzed in Dearden et al. (2024) “3D models related to the publication: The pharynx of the iconic stem-group chondrichthyan Acanthodes Agassiz, 1833 revisited with micro computed tomography.” Zoological Journal of the Linnean Society
Acanthodes confusus MNHN-F-SAA20 View specimen

|
M3#14703D surfaces representing the three-dimensionally fossilised head of Acanthodes confusus Type: "3D_surfaces"doi: 10.18563/m3.sf.1470 state:published |
Download 3D surface file |
Acanthodes confusus MNHN-F-SAA21 View specimen

|
M3#14713D surfaces representing the three-dimensionally fossilised head of Acanthodes confusus Type: "3D_surfaces"doi: 10.18563/m3.sf.1471 state:published |
Download 3D surface file |
Acanthodes confusus MNHN-F-SAA24 View specimen

|
M3#14723D surfaces representing the three-dimensionally fossilised head of Acanthodes confusus Type: "3D_surfaces"doi: 10.18563/m3.sf.1472 state:published |
Download 3D surface file |
This contribution contains the 3D models described and figured in the following publication: Georgalis, G.L., K.T. Smith, L. Marivaux, A. Herrel, E.M. Essid, H.K. Ammar, W. Marzougui, R. Temani and R. Tabuce. 2024. The world’s largest worm lizard: a new giant trogonophid (Squamata: Amphisbaenia) with extreme dental adaptations from the Eocene of Chambi, Tunisia. Zoological Journal of the Linnean Society. https://doi.org/10.1093/zoolinnean/zlae133
Terastiodontosaurus marcelosanchezi ONM CBI-1-645 View specimen

|
M3#1561Holotype maxilla ONM CBI-1-645 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1561 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-646 View specimen

|
M3#1560Paratype dentary ONM CBI-1-646 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1560 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-648 View specimen
|
M3#1562Maxilla ONM CBI-1-648 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1562 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-649 View specimen

|
M3#1559Maxilla ONM CBI-1-649 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1559 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-650 View specimen

|
M3#1563Maxilla ONM CBI-1-650 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1563 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-651 View specimen

|
M3#1564Maxilla ONM CBI-1-651 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1564 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-653 View specimen

|
M3#1565Maxilla ONM CBI-1-653 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1565 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-654 View specimen

|
M3#1576Maxilla ONM CBI-1-654 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1576 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-657 View specimen

|
M3#1566Dentary ONM CBI-1-657 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1566 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-658 View specimen

|
M3#1567Premaxilla ONM CBI-1-658 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1567 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-659 View specimen

|
M3#1568Dentary ONM CBI-1-659 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1568 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-660 View specimen

|
M3#1569Dentary ONM CBI-1-660 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1569 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-661 View specimen

|
M3#1570Dentary ONM CBI-1-661 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1570 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-668 View specimen

|
M3#1571Dentary ONM CBI-1-668 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1571 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-670 View specimen

|
M3#1572Dentary ONM CBI-1-670 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1572 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-672 View specimen

|
M3#1573Premaxilla ONM CBI-1-672 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1573 state:published |
Download 3D surface file |
Terastiodontosaurus marcelosanchezi ONM CBI-1-711 View specimen

|
M3#1574Premaxilla ONM CBI-1-711 of Terastiodontosaurus marcelosanchezi from the Eocene of Chambi Type: "3D_surfaces"doi: 10.18563/m3.sf.1574 state:published |
Download 3D surface file |
Todrasaurus gheerbranti UM THR 407 View specimen

|
M3#1575Holotype dentary UM THR 407 of Todrasaurus gheerbranti Type: "3D_surfaces"doi: 10.18563/m3.sf.1575 state:published |
Download 3D surface file |

The 3D dataset presented in this article provides the 3D models of two Chelonioidea turtles dentaries from the Paleocene of France described in: Lapparent de Broin F. de, Marek H., Barrier P. & Gagnaison C. 2025. — Euclastidae n. fam. (Chelonioidea) et première mention d’Euclastes Cope, 1867 dans le Paléocène du bassin de Paris (France). Geodiversitas 47 (10): 409-464. https://doi.org/10.5252/geodiversitas2025v47a10.
Euclastes wielandi ULB-04A21-10 View specimen

|
M3#1791Euclastes wielandi Type: "3D_surfaces"doi: 10.18563/m3.sf.1791 state:published |
Download 3D surface file |
Euclastes wielandi MNHN.F.BPT52 View specimen

|
M3#1709Euclastes wielandi (cast) Type: "3D_surfaces"doi: 10.18563/m3.sf.1709 state:published |
Download 3D surface file |
Euclastes wielandi ULB-04A21-11 View specimen

|
M3#1792Euclastes montenati nov. sp. Type: "3D_surfaces"doi: 10.18563/m3.sf.1792 state:published |
Download 3D surface file |
Euclastes wielandi MNHN.F.BPT53 View specimen

|
M3#1711Euclastes montenati (cast) Type: "3D_surfaces"doi: 10.18563/m3.sf.1711 state:published |
Download 3D surface file |
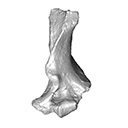
This contribution contains the 3D model described and figured in the following publication: Crochet, J.-Y., Hautier, L., Lehmann, T., 2015. A pangolin (Manidae, Pholidota, Mammalia) from the French Quercy phosphorites (Pech du Fraysse, Saint-Projet, Tarn-et-Garonne, late Oligocene, MP 28). Palaeovertebrata 39(2)-e4. doi: 10.18563/pv.39.2.e4
Necromanis franconica UM PFY 4051 View specimen
|
M3#12A partial left humerus from Pech du Fraysse (Saint-Projet, Tarn-et-Garonne, France), MP 28 (late Oligocene) Type: "3D_surfaces"doi: 10.18563/m3.sf12 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models analyzed in Neogene sloth assemblages (Mammalia, Pilosa) of the Cocinetas Basin (La Guajira, Colombia): implications for the Great American Biotic Interchange. Palaeontology. doi: 10.1111/pala.12244
cf. Nothrotherium indet. MUN STRI 12924 View specimen

|
M3#106Fragmentary basicranium with posterior portion of the skull roof. Type: "3D_surfaces"doi: 10.18563/m3.sf.106 state:published |
Download 3D surface file |
indet. indet. MUN STRI 16535 View specimen

|
M3#107Complete left ulna of a Scelidotheriinae gen. et sp. indet. Type: "3D_surfaces"doi: 10.18563/m3.sf.107 state:published |
Download 3D surface file |

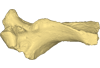
The presented dataset contains the 3D surface scan of the holotype of Birgeria americana, a partial skull described and depicted in: Romano, C., Jenks, J.F., Jattiot, R., Scheyer, T.M., Bylund, K.G. & Bucher, H. 2017. Marine Early Triassic Actinopterygii from Elko County (Nevada, USA): implications for the Smithian equatorial vertebrate eclipse. Journal of Paleontology. https://doi.org/10.1017/jpa.2017.36 .
Birgeria americana NMMNH P-66225 View specimen

|
M3#175NMMNH P-66225 is from upper lower Smithian to lower upper Smithian beds (Thaynes Group). The collecting site is located about 2.75 km south-southeast of the Winecup Ranch, east-central Elko County, Nevada, USA. P-66225 is a partial skull preserved within a large limestone nodule, with its right side exposed. It preserves the portion between the cleithrum posteriorly, and the level of the hind margin of the orbital opening anteriorly. The fossil has a length of 26 cm. Type: "3D_surfaces"doi: 10.18563/m3.sf.175 state:published |
Download 3D surface file |
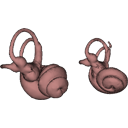
The present 3D Dataset contains the 3D models analyzed in Velazco P. M., Grohé C. 2017. Comparative anatomy of the bony labyrinth of the bats Platalina genovensium (Phyllostomidae, Lonchophyllinae) and Tomopeas ravus (Molossidae, Tomopeatinae). Biotempo 14(2).
Platalina genovensium 278520 View specimen

|
M3#276Right bony labyrinth surface positioned (.PLY) Labels associated (.FLG) Type: "3D_surfaces"doi: 10.18563/m3.sf.276 state:published |
Download 3D surface file |
Tomopeas ravus 278525 View specimen

|
M3#277Right bony labyrinth surface (.PLY) Labels associated (.FLG) Type: "3D_surfaces"doi: 10.18563/m3.sf.277 state:published |
Download 3D surface file |

This contribution contains the 3D models described and figured in the following publication: Tissier et al. (in prep.).
Sellamynodon zimborensis UBB MPS 15795 View specimen

|
M3#297Incomplete skull with left M3. Type: "3D_surfaces"doi: 10.18563/m3.sf.297 state:published |
Download 3D surface file |
Sellamynodon zimborensis UBB MPS 15795 View specimen

|
M3#298Mandible with complete molar and premolar rows, lacking symphysis. Type: "3D_surfaces"doi: 10.18563/m3.sf.298 state:published |
Download 3D surface file |
Amynodontopsis aff. bodei UBB MPS V545 View specimen

|
M3#299Maxillary fragment with M1-3. Type: "3D_surfaces"doi: 10.18563/m3.sf.299 state:published |
Download 3D surface file |
Amynodontopsis aff. bodei UBB MPS V546 View specimen

|
M3#300Unworn m1/2 on mandible fragment. Type: "3D_surfaces"doi: 10.18563/m3.sf.300 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models analyzed in the publication ‘Ontogenetic development of the otic region in the new model organism, Leucoraja erinacea (Chondrichthyes; Rajidae)’, https://doi.org/10.1017/S1755691018000993
Leucoraja erinacea 2018.9.26.1 View specimen

|
M3#3673D model of the right skeletal labyrinth of the adult specimen of Leucoraja erincea. T Type: "3D_surfaces"doi: 10.18563/m3.sf.367 state:published |
Download 3D surface file |
Leucoraja erinacea 2018.9.25.2 View specimen

|
M3#3683D model of the right skeletal labyrinth of the stage 34 specimen of Leucoraja erincea. Type: "3D_surfaces"doi: 10.18563/m3.sf.368 state:published |
Download 3D surface file |
Leucoraja erinacea 2018.9.25.3 View specimen

|
M3#3693D model of the right skeletal labyrinth of the stage 32 specimen of Leucoraja erinacea. Type: "3D_surfaces"doi: 10.18563/m3.sf.369 state:published |
Download 3D surface file |
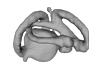
|
M3#3723D model of the right membranous system of stage 32 of Leucoraja erincea. Type: "3D_surfaces"doi: 10.18563/m3.sf.372 state:published |
Download 3D surface file |
Leucoraja erinacea 2018.9.25.4 View specimen

|
M3#3703D model of the right skeletal labyrinth of the stage 31 specimen of Leucoraja erinacea. Type: "3D_surfaces"doi: 10.18563/m3.sf.370 state:published |
Download 3D surface file |
Leucoraja erinacea 2018.9.26.5 View specimen

|
M3#3763D model of the right skeletal labyrinth of the stage 29 specimen of Leucoraja erinacea. Type: "3D_surfaces"doi: 10.18563/m3.sf.376 state:published |
Download 3D surface file |
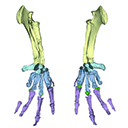
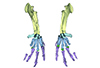
The present 3D Dataset contains the 3D model analyzed in Vautrin et al. (2019), Palaeontology, From limb to fin: an Eocene protocetid forelimb from Senegal sheds new light on the early locomotor evolution of early cetaceans.
?Carolinacetus indet. SNTB 2011-01 View specimen
|
M3#3983D model of an articulated forelimb of a Carolinacetus-like protocetid from Senegal Type: "3D_surfaces"doi: 10.18563/m3.sf.398 state:published |
Download 3D surface file |